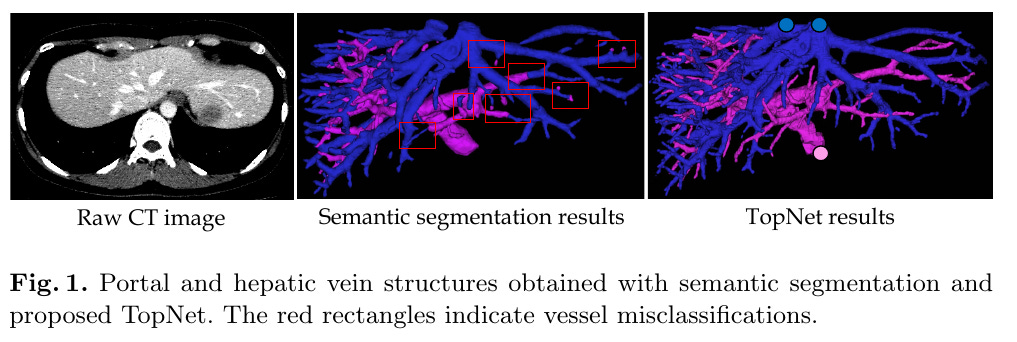
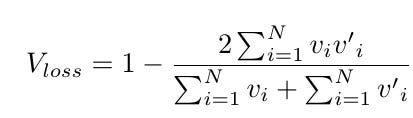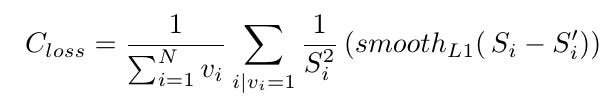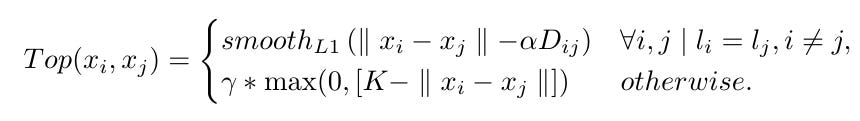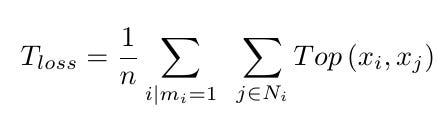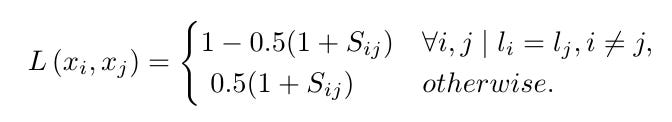TopNet: Topology Preserving Metric Learning
These are some of my notes for the MICCAI 2020 paper TopNet: Topology Preserving Metric Learning.
Here is the paper link.
Motivation
Reconstructing Portal Vein and Hepatic Vein trees from abdominal CT scans is a prerequisite for liver surgery.
Locally, both types of veins look similar, so dense segmentation based on patch-wise DL wouldn’t work, and the trees need to be traced to their source.
Proposed Solution: Learn to predict centerlines and a metric that connects these centerlines. Use source vessel detection to assign labels to vascular trees.
Introduction
Normal segmentation makes local mistakes which are amplifies the error across the image.
Contributions:
Multi-task network for tree reconstruction which detects centerlines and connectivity between centerlines.
Topology metric which learns both intra and interclass topological distance between vascular voxel pairs.
Verified better accuracy
Method
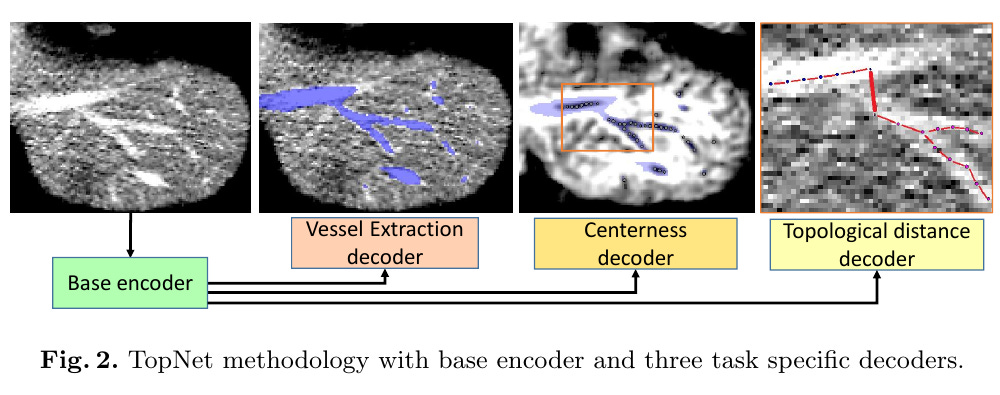
There are 3 parts in their multi-task network.
1. Vessel extraction decoder
Separate vessels from background. Formulated as a semantic segmentation problem. Use dice loss to train this task.
where v is the ground truth binary vessel volume patch, v′ is the predicted volume patch.
2. Centerness decoder
The centerness score can be formulated as a regression problem (it will be unstable to train as a binary segmentation problem).
Let mi∈M be the binary volume representing center voxels.
For a given voxel j, define distance transform:
The values are considered only for points inside vessel (a lot of points outside vessel will skew the output otherwise).
Use smooth L1 loss:
The denominator ensures that the outputs are not skewed towards voxels away from center (there are ∝S2 voxels at a distance S from centerline).
Perform NMS (non-maximal suppression) over a window size of 5×5×5.
3. Topology distance Decoder
This network outputs a 8d feature vector for each voxel in CT volume.
The idea here is that the distance between the features xi,xj at voxels i,j given by ‖xi−xj‖ corresponds to the topological distance in the vessel space if they are in the same tree.
If they are in different trees, the distance should be large.
Given labels li,lj for voxels i,j the loss function is given by:
For a given input image patch, the loss is:
where Ni is a neighborhood of voxel ii.
The neighborhood is defined inside a radius of 15 voxels ⇒ topological distance is bounded.
They set α=115 to approximately normalize topological distance αDij from 0 to 1. Also set K=3. To balance loss terms, set γ=13.
Tree reconstruction (during test-time post-training)
Use vessel prediction to mask center-line distance.
Use NMS on center-line distance Si to find center voxels.
For voxel i, create edges for all nodes j where ‖xi−xj‖≤2.
Use Dijkstra’s multi-source shortest path tree algorithm.
Experiments
Datasets
IRCAD public dataset, internal dataset.
Methods
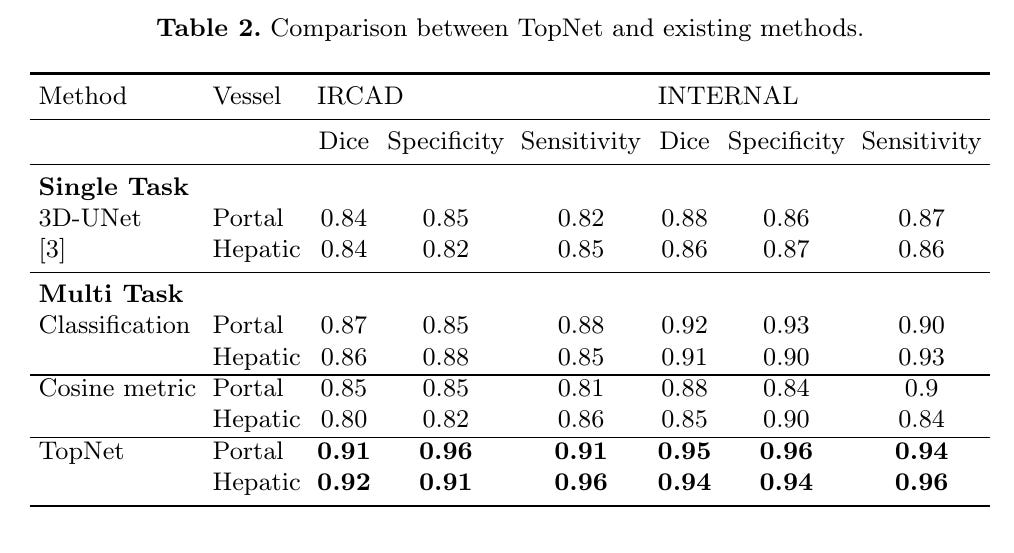
Comparison with single task 3D UNet (dice loss).
For multi-task, replace topology distance with 2-channel probability map (dice loss over all center-voxels).
Multi-task cosine metric learning (Sij = cosine distance)
For tree reconstruction, check where Sij<=0.5 and take edge weights as:
wij=Eij(1−Sij)
where Eij is the euclidean distance between voxels i,j.
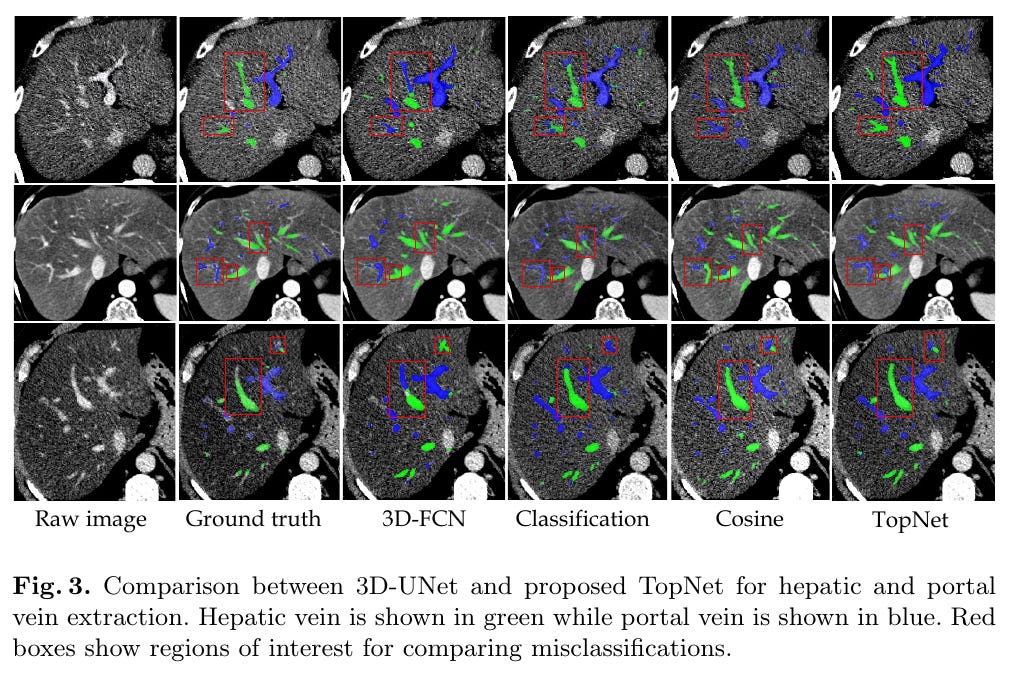
Without metric learning, there is abrupt change in vessel labels along vascular tree.
Probably reason is that cosine based or single-task try to classify based on frequently observed vessel patterns.
But topology metric explicitly constrains to tracing vessels (so that the topology distance is correct)